Latest news
 1st Network Meeting
1st Network Meeting
November 15th, 2021
 ENLIGHT-TEN+ was virtually kicked-off on March 4th, 2021
ENLIGHT-TEN+ was virtually kicked-off on March 4th, 2021
 Start of ENLIGHT-TEN+
Start of ENLIGHT-TEN+
March 1st, 2021
ENLIGHT-TEN+
A structured research & training network of European immunologists and bioinformaticians from academia and industry focused on the in-depth characterisation and tailored targeting of tissue-resident T cells
ENLIGHT-TEN+ is a European Network Linking Informatics and Genomics of Helper T cells in Tissues. This Innovative Training Network (ITN), which is based on the successfully conducted ITN ENLIGHT-TEN (2015-2019) and funded by the European Union H2020 Programme, addresses a crucial missing link in our current research environment, the lack of qualified individuals who possess the cellular immunology skills to recognize and define important scientific questions amenable to -omics approaches, and also the bioinformatic expertise to interrogate and interpret the resulting big-data appropriately. Thus, ENLIGHT-TEN+’s mission is to provide cross-disciplinary training for a new generation of enthusiastic researchers who have in-depth understanding of T cell immunology and are also capable to handle large datasets. The network of 15 beneficiaries and 8 partner organisations from 10 European countries combines ample expertise on T cell biology with state-of-the-art technologies generating big-data, cutting-edge bioinformatic tools including artificial intelligence, preclinical models and samples from patient cohorts. ENLIGHT-TEN+ will integrate highly innovative -omics-based technologies to generate a novel view of tissue-resident T cells, understand the microenvironmental cues & molecular factors shaping their unique functional properties with unprecedented resolution, and apply this understanding to immune-mediated pathologies, allowing us to selectively manipulate tissue-resident T cells in autoimmune diseases. ENLIGHT-TEN+ offers an outstanding programme of scientific education and doctoral training for 15 early-stage researchers (ESRs) across Europe. These ESRs will be empowered to push our knowledge of tissue-resident T cells beyond the state-of-the-art, which will enable the identification of novel biomarkers and support the development of advanced therapeutic concepts through close collaboration between academic and industrial partners.
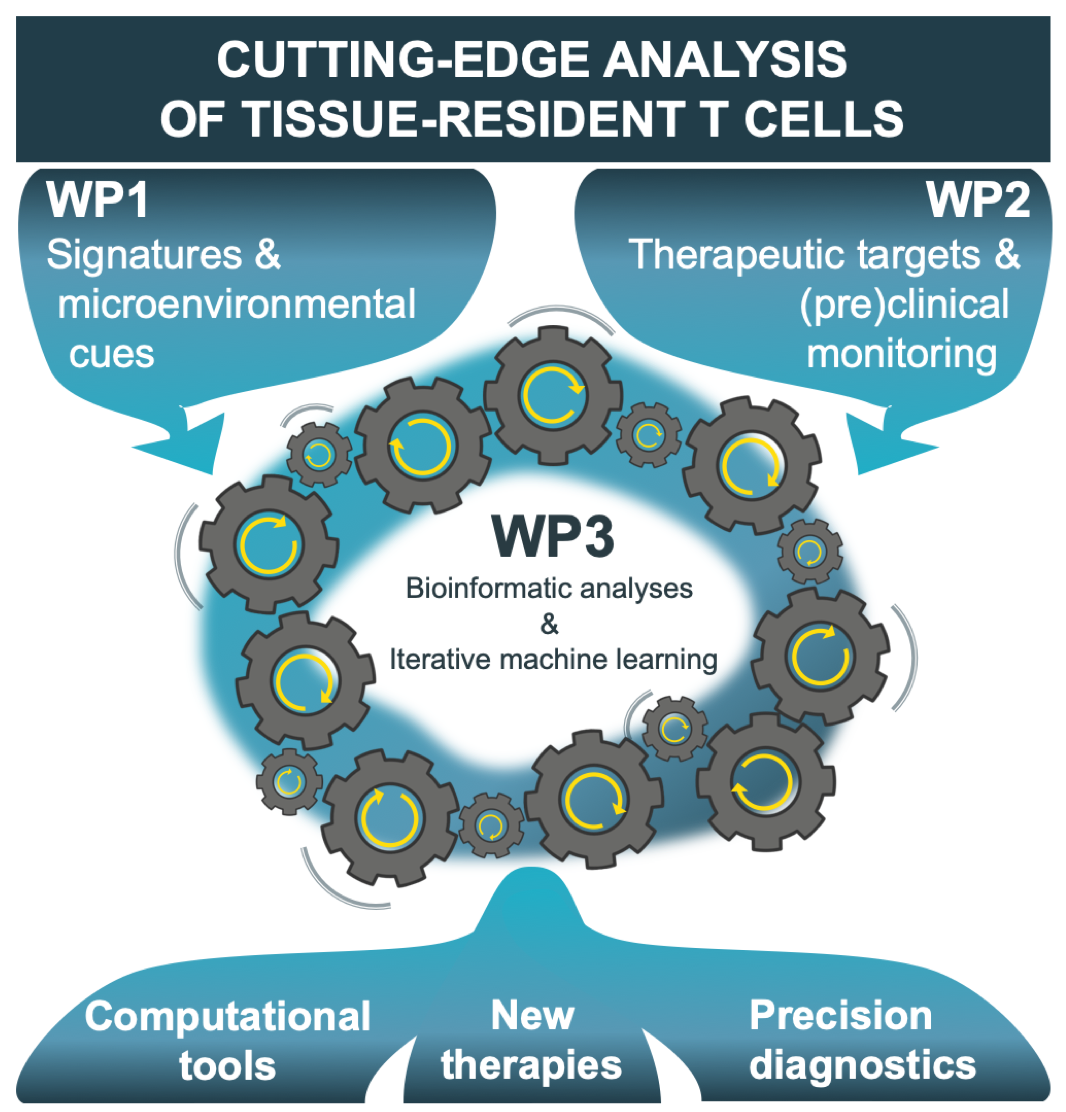
Integration of ENLIGHT‑TEN+’s scientific work packages (WPs)
The generation of big-data is an emerging and challenging field, and there is high demand for researchers to be able to analyse, integrate and exploit this rich source of information in order to deliver new scientific understanding, new technologies and new treatments. ENLIGHT-TEN+ will combine individual strengths of innovative laboratories and enterprises from complementary disciplines to provide unique intersectoral, interdisciplinary & international training as an ideal stepping-stone for the ESRs to enter and strengthen Europe’s academia as well as pharmaceutical and bioinformatics companies.
Autoimmune diseases arise as a consequence of aberrant immune responses against the body’s own tissues. The clinical symptoms and nature of the autoimmune diseases vary according to the tissue being targeted – e.g. gut, pancreas, liver, joints, central nervous system – but these diseases are all underpinned by a common etiology: a dysregulated immune system. There are more than 80 different types of autoimmune diseases, and their prevalence is increasing. Many autoimmune diseases are driven by CD4+ helper T cell responses, and recent findings suggest that chronicity of autoimmune diseases largely depends on the reactivation of tissue-resident memory T (TRM) cells1, 2 that reside in almost all tissues of the body. Therefore, precision diagnostics & therapeutic strategies that target TRM cells have emerged as key priorities in the quest to tackle the scourge of autoimmune diseases.
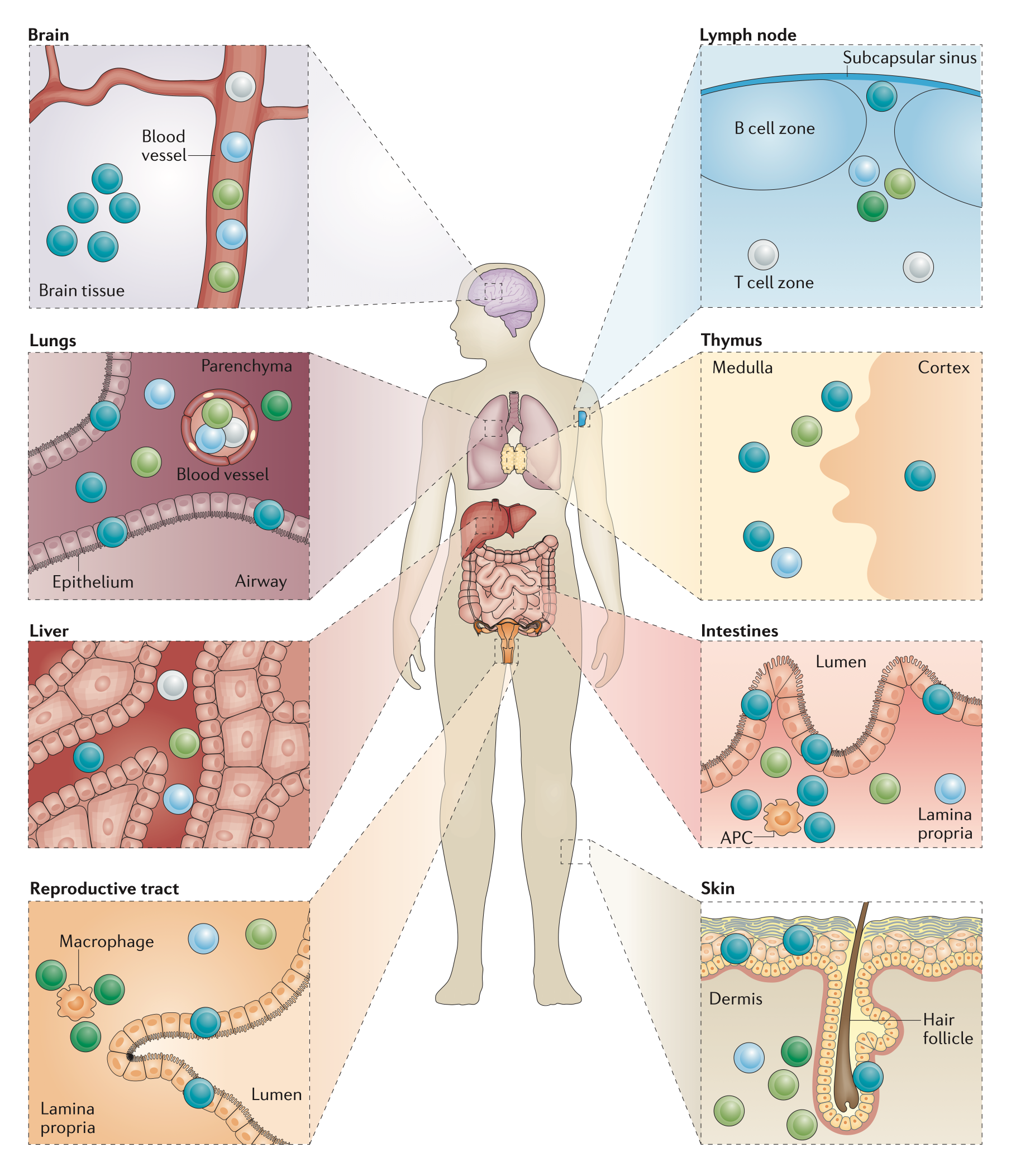
Tissue-resident T cells. Memory T cell subsets have a distinct microanatomical localization
within non-lymphoid tissues (modified from Mueller&Mackay Nat Rev Immunol 16, 79).
TRM cells are strongly influenced by the ‘conditioning’ in the tissue. In healthy settings, this conditioning tailors the immune response to the precise need for organ-specific defence, however, in patients with chronic autoimmune diseases, environmental conditioning may modulate an ongoing tissue-resident memory response to unexpectedly trigger pathology. Very often, the molecular factors controlling the tissue-specific conditioning as well as reactivation of TRM cells in the context of autoimmunity are incompletely understood, as the interaction of the TRM cells with their microenvironment is extremely complex. As a consequence, there is a pressing need to apply cellular immunology & -omics approaches to TRM cells derived from steady-state and autoimmune conditions in order to better understand their identity and function3 and to fully harness their therapeutic potential.
In parallel with the emergence of the tissue-residency paradigm, the world is witnessing a big-data revolution. The power of big-data is percolating every aspect of the European society, and the field of immune-mediated diseases is no exception. Understanding how aberrant behaviour of immune cells leads to disease pathology can be greatly expedited by harnessing the power of technologies that yield high-dimensional data. Big-data approaches are particularly applicable to the study of T cells because of the high degree of complexity associated with their differentiation, plasticity, memory capability and adaptation to environmental niches. While refined techniques have emerged for the efficient and cost-effective generation of various -omics and high-dimensional flow cytometry data, the capability to extract biologically meaningful information from such data has lagged behind. The big-data revolution is now paving the way for these large datasets to be interrogated and interpreted in novel ways, permitting the generation of new insights that will be critical for understanding and treating T cell-mediated autoimmune diseases.
Recognizing the urgent need for a new type of researcher that is truly at home with both wet-lab and computational approaches, leading scientists in the field founded ENLIGHT-TEN+. Addressing key immunological questions is too complex for mono-dimensional thinking and education. Instead it needs the exchange across disciplines & sectors to creatively and jointly tackle existing problems as well as emerging challenges. At the end of the project, the 15 ESRs of ENLIGHT TEN+ will be well prepared for their professional career, as they fill the existing gulf between wet-lab and computational disciplines, thereby significantly increasing their employability.
References
1 Masopust&Soerens (2019) Tissue-resident T cells and other resident leukocytes. Annu Rev Immunol 37, 5212 Wu et al. (2018) Pathogenic role of tissue-resident memory T cells in autoimmune diseases. Autoimmun Rev 17, 906
3 Mackay&Kallies (2017) Transcriptional regulation of tissue-resident lymphocytes. Trends Immunol 38, 94